Microbead-based microfluidics (droplet-based microfluidic using specific gel as dispersed phase) is a powerful technic which…

Customer paper on biomolecular circuits with droplets
Cell-free extract based optimization of biomolecular circuits with droplet microfluidics
By Yutaka Hori, Chaitanya Kantak, Richard M. Murray and Adam R. Abate from the UCSFand CalTech in the USA.
Engineering an efficient biomolecular circuit often requires timeconsuming iterations of optimization. Cell-free protein expression systems allow rapid testing of biocircuits in vitro, speeding the design–build–test cycle of synthetic biology. In this paper, we combine this with droplet microfluidics to densely scan a transcription–translation biocircuit space. Our system assays millions of parameter combinations per hour, providing a detailed map of function. The ability to comprehensively map biocircuit parameter spaces allows accurate modeling to predict circuit function and identify optimal circuits and conditions.
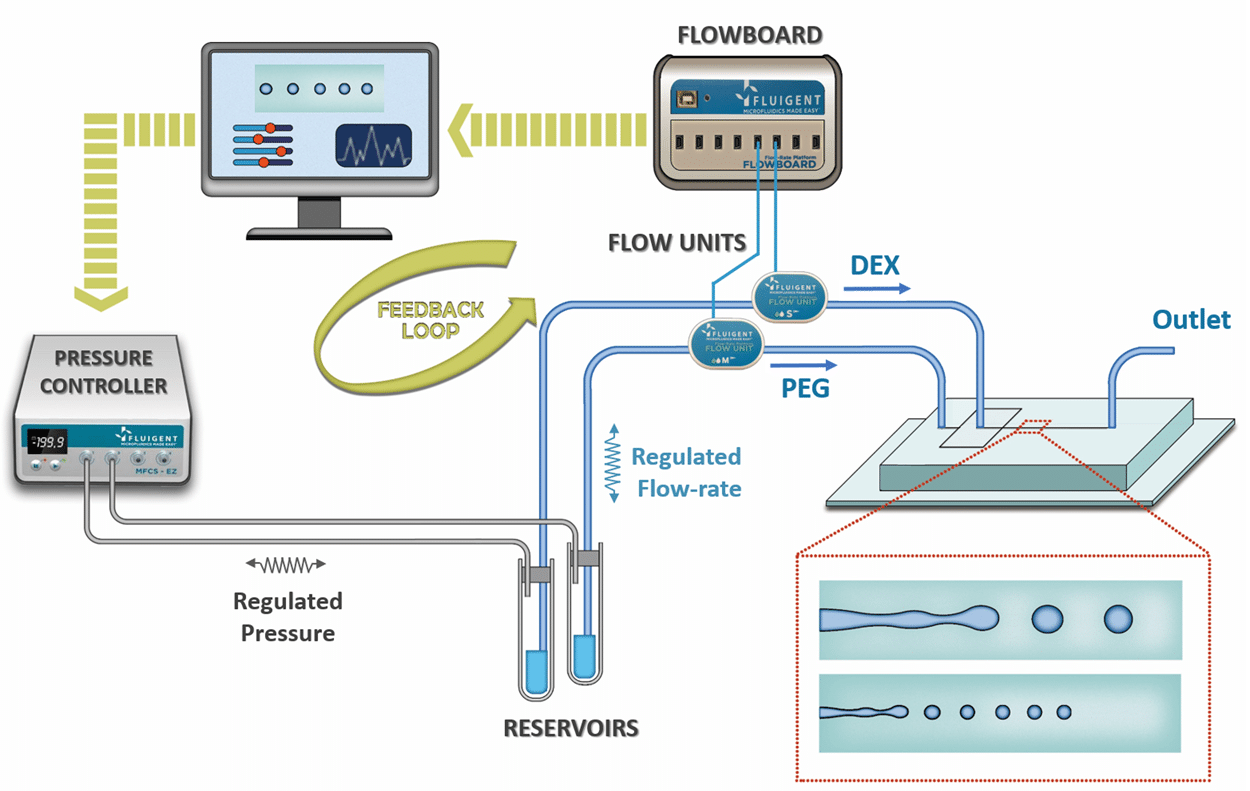
This was made possible with the use our pressure-driven flow controller pump system (MFCS™-EZ) which allows independent control of each inlet, enabling droplet composition to be modulated by dynamically varying flow rates during droplet generation.
For more information: doi:10.1039/C7LC00552K or Lab Chip, 2017,17, 3037-3042.