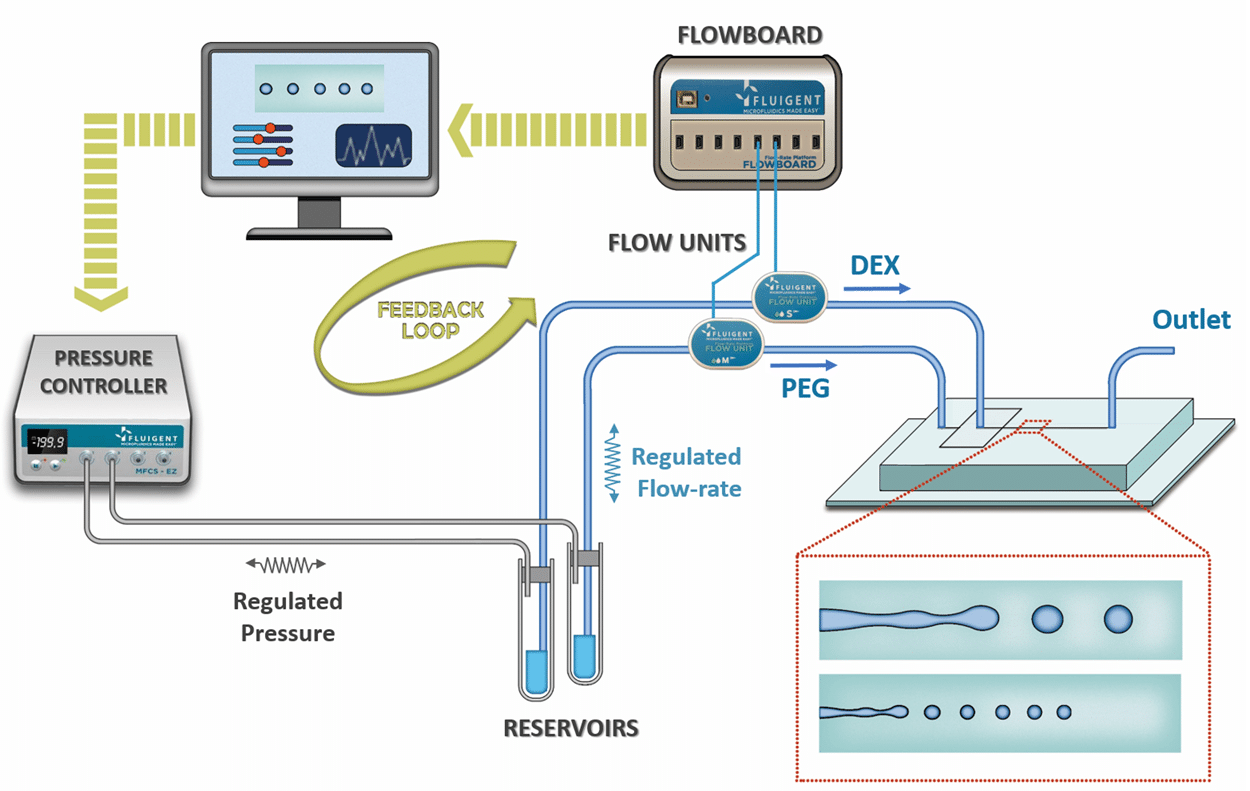
Microbead-based microfluidics (droplet-based microfluidic using specific gel as dispersed phase) is a powerful technic which…

New publication from Japan and France on biochemical circuits
High-resolution mapping of bifurcations in nonlinear biochemical circuits
By A. J. Genot, A. Baccouche, R. Sieskind, N. Aubert-Kato, N. Bredeche, J. F. Bartolo, V. Taly, T. Fujii and Y. Rondelez, from French and Japan Universities.
“Analog molecular circuits can exploit the nonlinear nature of biochemical reaction networks to compute low-precision outputs with fewer resources than digital circuits. This analog computation is similar to that employed by gene-regulation networks. Although digital systems have a tractable link between structure and function, the nonlinear and continuous nature of analog circuits yields an intricate functional landscape, which makes their design counter-intuitive, their characterization laborious and their analysis delicate. Here, using droplet-based microfluidics, we map with high resolution and dimensionality the bifurcation diagrams of two synthetic, out-of-equilibrium and nonlinear programs: a bistable DNA switch and a predator–prey DNA oscillator. The diagrams delineate where function is optimal, dynamics bifurcates and models fail. Inverse problem solving on these large-scale data sets indicates interference from enzymatic coupling. Additionally, data mining exposes the presence of rare, stochastically bursting oscillators near deterministic bifurcations.”
These experiments were made possible with the use our pressure-driven flow controller pumps.
For more information: doi:10.1038/nchem.2544 or Nature Chemistry, 8, 760–767 (2016)